Detection of pathogens and cancer biomarkers utilizing custom designed DNA-binding proteins and investigation of the engineered protein-DNA interactions
- Detection of pathogens
- Detection of cancer biomarkers
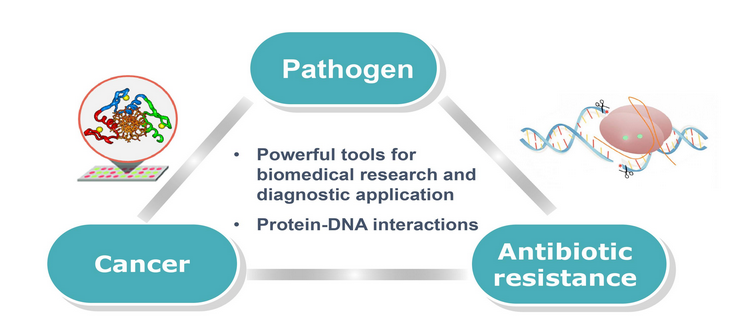
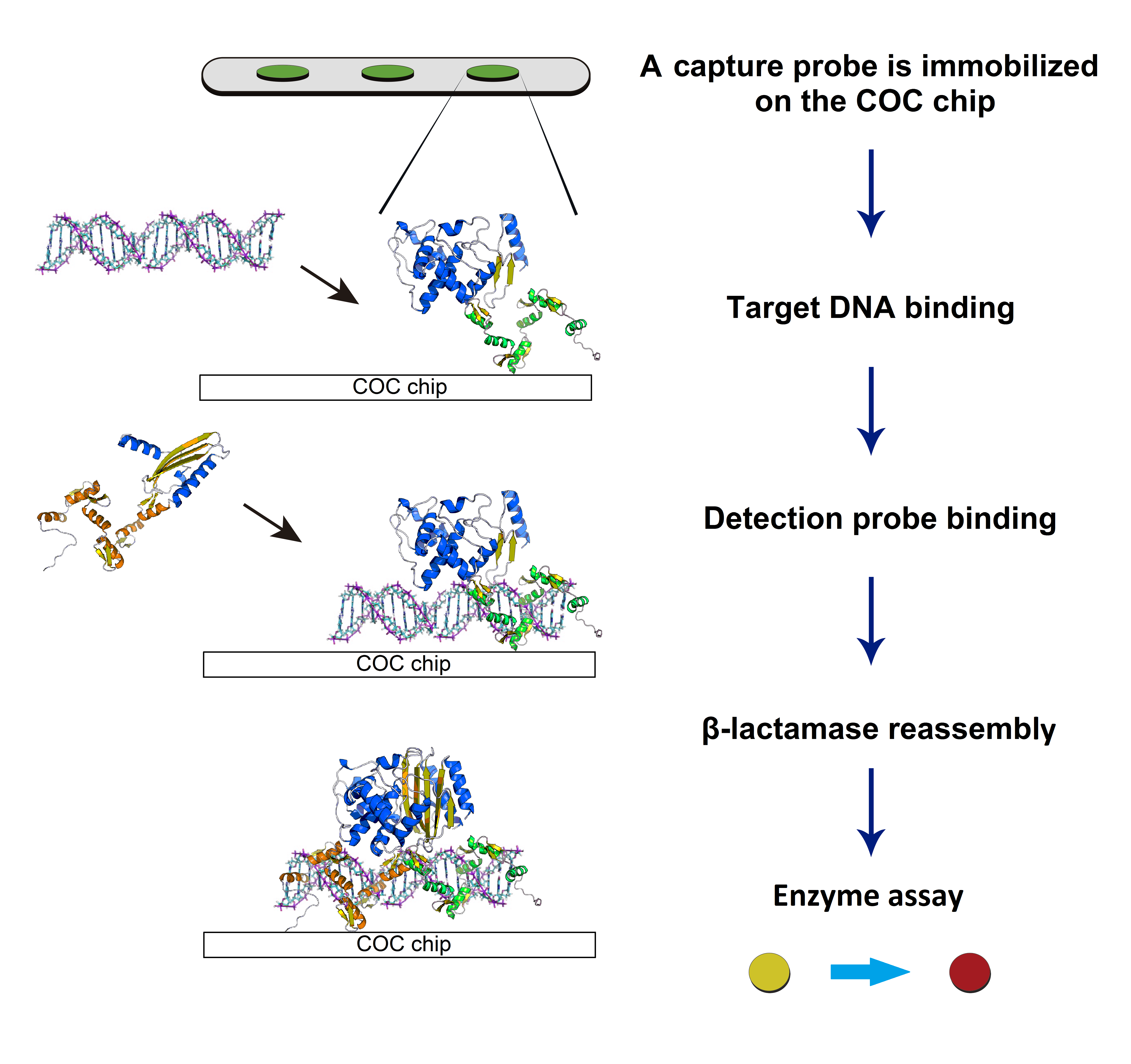
The sensitive detection of DNA is of great interest and demand for an increasing number of biomedical studies and diagnostic applications. The development of simple, rapid, and robust technologies for detection of pathogens is becoming particularly critical, considering the increasing rate of mortality and risk of bacteremia. The use of DNA-binding proteins provides a useful scaffold for ‘direct’ reading of the sequence information, avoiding the need for denaturation and subsequent renaturation required by the DNA hybridization method. Thus, DNA-binding domains such as zinc fingers (ZFs) and novel transcriptional activator-like effectors (TALEs) are engineered for recognition of specific sequences present in multiple pathogens. We also probe DNA recognition and binding behaviors of engineered TALE proteins using a combined biochemical, biophysical, and computational (bioinformatics) approach to provide a new insight into protein-DNA interactions. We envision a protein array capable of multiplexed detection of various pathogens. Our effort to incorporate a molecular diagnostic on a microfluidic technology could possibly facilitate the development of a point-of-care device.
We also plan to explore the potential use of our system for the detection of specific sites of DNA methylation linked to cancer. Current methylated CpG detection methods are PCR-based and involve the bisulfate modification of DNA, followed by DNA sequencing or methylation-specific PCR. These methods require extensive bisulfate reaction times (4-18 hrs) along with false positive problems caused by incomplete bisulfate reactions. The availability of a simple molecular method to directly determine CpG methylation in a promoter-specific manner would provide a powerful method for the detection of a specific cancer. Combined with the methyl-CpG binding domains, our system will be further developed to detect promoter hypermethylation of tumor suppressor genes in urine DNA sample from bladder cancer patients. Detection of aberrant methylation of candidate genes with our system can provide a non-invasive method for bladder cancer detection by obviating the need for a procedure to obtain biopsy material. It can also serve as early diagnosis and prognosis for bladder cancer.
Novel application of 2D-nanosheet and DNA-binding proteins for rapid detection of antibiotic resistance genes/bacteria.
DNA detection technologies play an important role in diagnostic applications in the areas of public health and biomedicine. There is a growing need to improve our capability in environmental and biomedical diagnostics. In particular, antibiotic resistance bacteria (ARB) has become a threatening health issue worldwide due to these bacterial infections associated with significant morbidity and mortality. Consequently, antibiotic resistance genes (ARGs) are thought to be emerging environmental contaminants. Thus, the development of a new class of simple, rapid, and robust technology for screening and detecting antibiotic resistance genes (ARGs) would be beneficial for rapidly tracking the sources and spread of ARGs in the environment, reducing potential health risks. The goal of this project is to develop a new sensing system for detecting multiple ARGs by employing nanosheets and DNA-binding proteins. We have developed a transient plant expression system using Nicotiana Benthamiana to express the custom-designed TALE proteins. We explore the potential of graphene oxide (GO) to develop a new sensing platform for direct detection of dsDNA.
Developing the CRISPR/dCas9-based DNA sensor using the graphene oxide (GO) as a new sensing platform
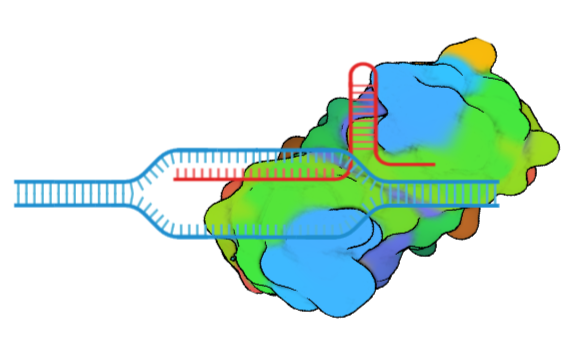
The CRISPR/Cas9 system is an emerging tool in genome engineering, consisting of a Cas9 endonuclease and a guide RNA which binds to a target in the DNA. In this study, CRISPR/dCas9 with single guided RNA (sgRNA) will detect specific double stranded DNA sequences in target gene. We are developing CRISPR/dcas9 system in combination with nanomaterials which can function as a rapid, facile, and reliable gene detection system with multiplexing capabilities.
Research Support
- NSF
- Kentucky Biomedical Research Infrastructure Network (KBRIN)-NIH INBRE
- NSF EPSCoR
- USDA/WKU
- WKU - ARTP Collaborative Research Grant(2013), RCAP(2015)


